Transform Your R Reports
scientific provides two highly customizable RMarkdown templates designed specifically for scientific research and technical documentation, including machine learning and AI reports.
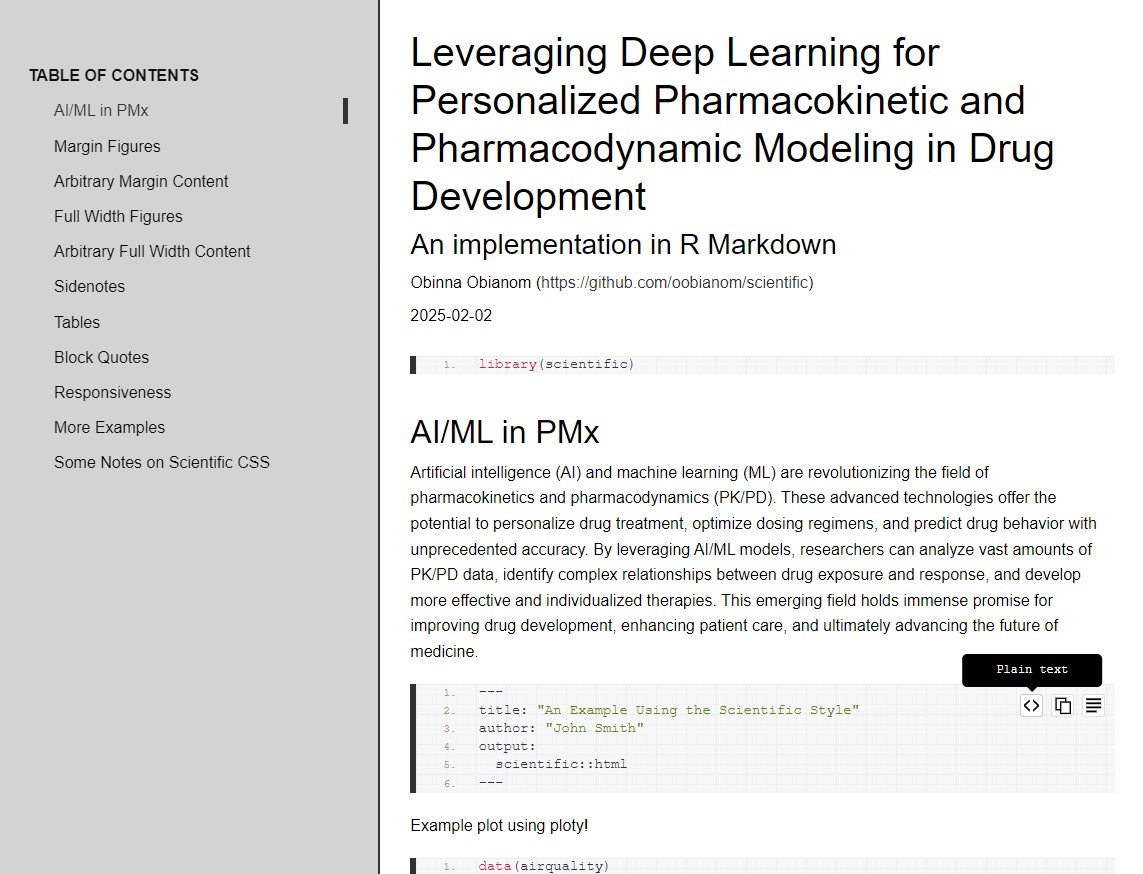
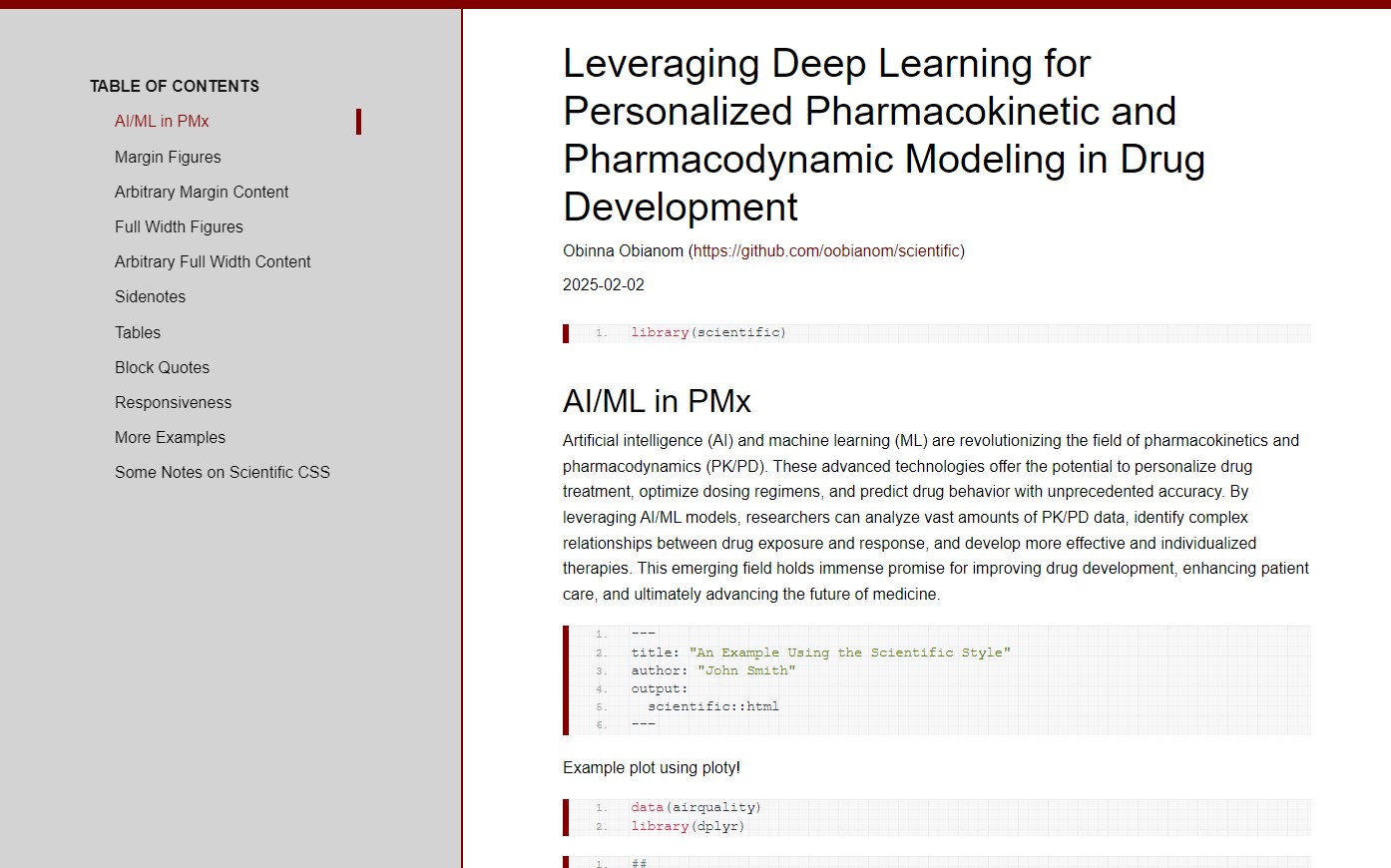
Template 1 screenshots

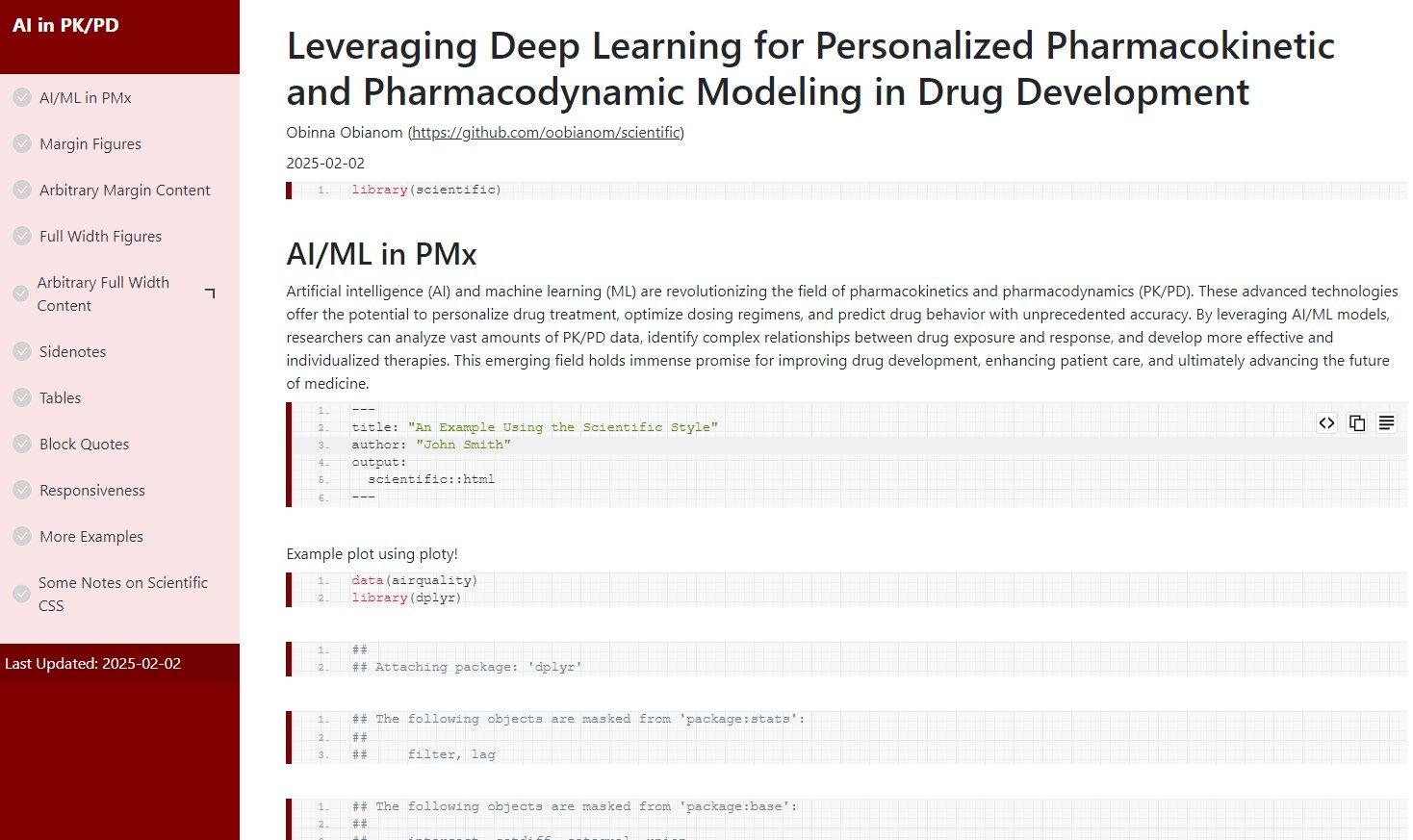
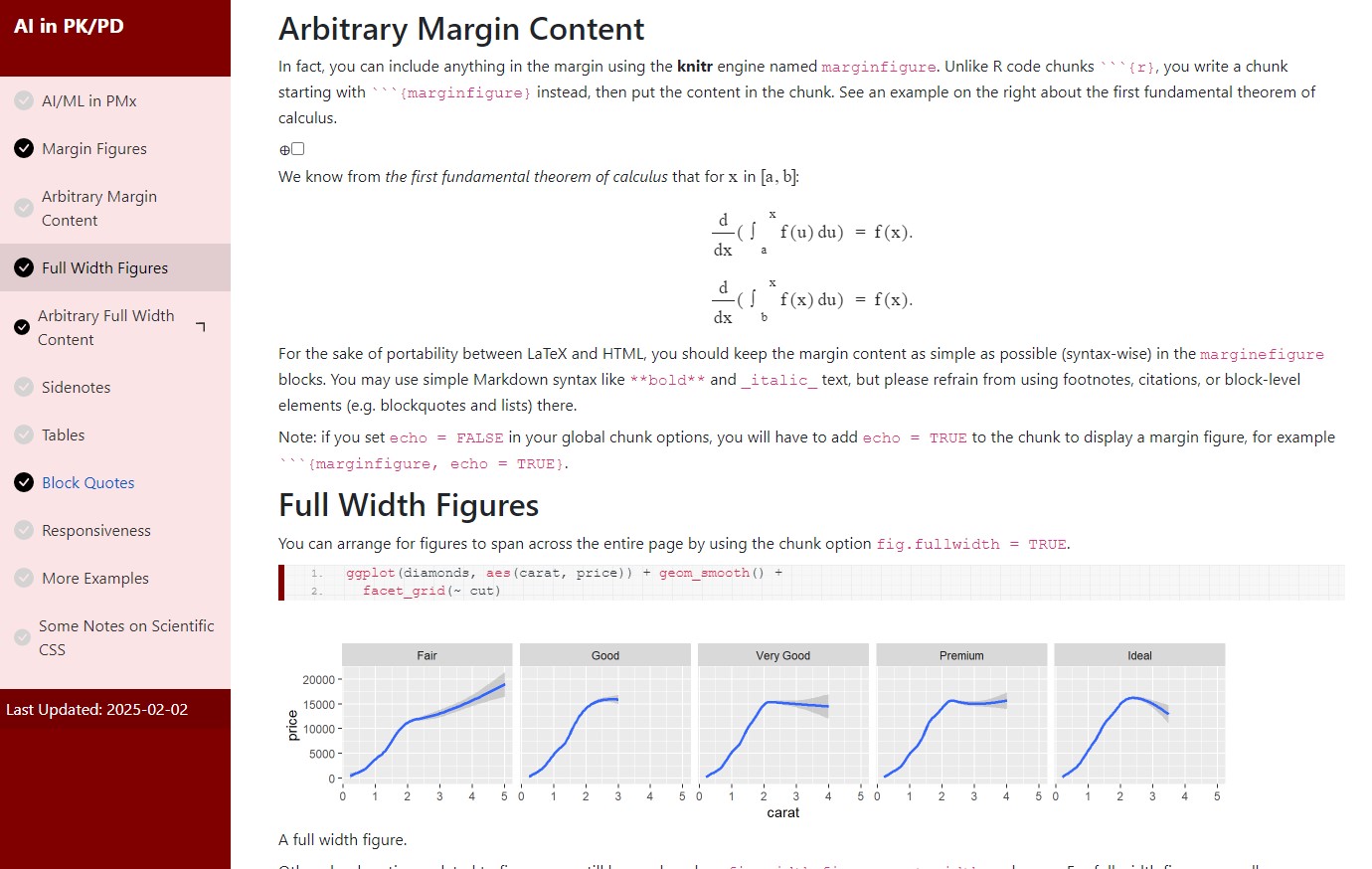
Template 2 screenshots

Quick R Installation
install.packages("scientific")
Usage .Rmd Example
title: "Scientific Report Example"
highlighter: "dracula" # code theme
codelang: "r" #default is R, you may add js php python and so on
themecolor: "#800000" #specify a theme color
author: "Obinna Obianom"
date: "`r Sys.Date()`"
output:
scientific::html:
toc: TRUE
self_contained: TRUE
template: template1 # or template2
summaryslide: TRUE # whether to show summary slide on top
lightsummaryslide: FALSE # whether to make summary slide light
summarypoints:
- Fusce id velit ut tortor pretium viverra. Lectus urna duis convallis convallis tellus id interdum.
- Erat imperdiet sed euismod nisi porta lorem. Sed viverra tellus in hac habitasse platea dictumst.
- Turpis egestas integer eget aliquet nibh praesent tristique. Ultricies integer quis auctor elit.
- Dictum varius duis at consectetur lorem donec massa. Convallis aenean et tortor at risus viverra.
- Egestas tellus rutrum tellus pellentesque eu tincidunt tortor aliquam nulla. Aliquet bibendum enim facilisis gravida neque.
- Porta lorem mollis aliquam ut porttitor leo a diam. Sit amet mattis vulputate enim nulla aliquet porttitor lacus.
- Vestibulum rhoncus est pellentesque elit ullamcorper dignissim. Risus feugiat in ante metus dictum.
---
# Introduction
Sample text implemented in LaTeX and HTML/CSS
```{r, message=FALSE}
library(ggplot2)
mtcars2 <- mtcars
mtcars2$am <- factor(
mtcars$am, labels = c('automatic', 'manual')
)
ggplot(mtcars2, aes(hp, mpg, color = am)) +
geom_point() + geom_smooth() +
theme(legend.position = 'bottom')
```
Some final texts